Darci Trader Lab
Menu
Research
Targeting the proteasome: Beyond Inhibition
|
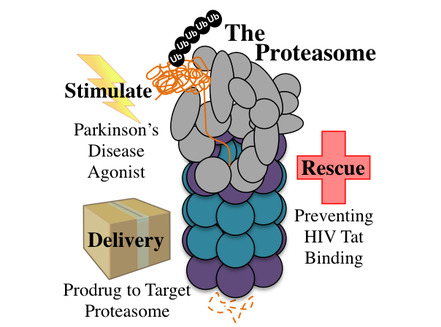
The proteasome is a multiprotein, multicatalytic site complex that acts as the main pathway for protein degradation in cells. My research program focuses on the development of chemical strategies to control and harness proteasome-mediated protein degradation. Unlike traditional work in this field that has focused on the discovery of proteasome inhibitors, my program will seek to identify compounds that enable us to stimulate, rescue and direct protein degradation. We will harness the power of the proteasome through pursuit of three synergistic projects: 1) chemical stimulation of the 20S proteasome in neurons to abate the protein aggregation events that lead to Parkinson’s disease, 2) rescue of the proteasome from inhibition by HIV Tat, which results in limited antigenic peptide production, substantially reducing the immune response to infected cells, and 3) generation of a set of rules for selective targeting of the immunoproteasome to release an antigenic peptide, marking diseased cells for destruction by the immune system.
|
Proteasome Stimulation:
|
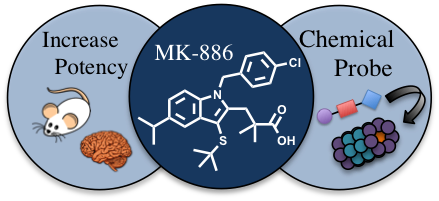
A number of neurological diseases can be attributed to the accumulation of a toxic protein. For example, in Parkinson’s disease (PD) alpha-synuclein, a small, unstructured protein normally present in low levels in neurons, is accumulated. It has been hypothesized that if the activity of the 20S proteasome can be selectively upregulated, this may result in a decrease in the level of alpha-synuclein, and diminish the rate of neuron decline in PD patients. During my postdoctoral tenure, I developed a method to screen for and validate small molecules that can act as 20S agonists. This strategy led to the identification of the agonist MK‑886. While this small molecule could penetrate neuron cells and decrease the amount of endogenously produced alpha-synuclein, its potency is poor (EC50≈ 32 µM). However, MK-886 provides a starting point for the design of more potent 20S agonists and development of probes to elucidate how this small molecule interacts with the 20S to stimulate protein degradation. The long-term goals of this work will be to determine the feasibility of using proteasome activation to treat PD and more broadly, the utility of such a strategy for the treatment of other protein accumulation-associated neurological diseases.
|
Rescuing the Proteasome:
Proteasome-Mediated Drug Delivery:
|
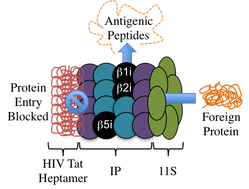
A number of viruses, including Hepatitis B, Epstein Barr, and the Human Immunodeficiency Virus (HIV) produce proteins that directly interact with the immunoproteasome (IP) to limit or stop protein proteolysis. The IP is similar in structure to the 20S proteasome, but the three catalytic moieties are exchanged with units that better produce antigenic peptides. “Rescue” of the IP from inhibition by virally-produced proteins would allow production of major histocompatibility complex molecules, class I (MHC I), which display non-self peptides to cytotoxic T-cells, triggering an immediate immune response. Specifically, we are interested in further studying how Tat, a small protein produced in an HIV infected cell, prevents the IP from generating peptides for MHC I display and development of a small molecule to prevent the IP‑HIV Tat interaction. Discovery and application of a peptidomimetic that inhibits binding of HIV Tat to the IP to enable MHC I production would be a welcomed addition to the arsenal of molecules to treat HIV.
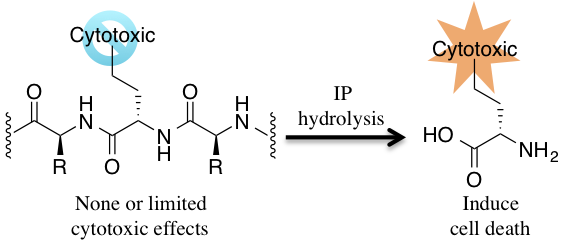
Diseased cells produce the IP in response to inflammation or viral infection and their selective IP expression can provide a mechanism to target diseased over healthy cells. Ideally, the IP facilitates production of antigenic peptides that are used to alert the immune system. However, this process is not always efficient which prevents the natural cellular machinery from initiating a sufficient immune response. I propose to study how the IP can be used to selectively release a “masked” antigenic peptide into diseased or infected cells that typically evade the immune system. Prodrug-like molecules containing an antigenic peptide will be designed such that this moiety will be released upon exposure to the IP, but not by the standard proteasome found in normal cells. Extensive studies will be required to understand the “rules” that dictate IP recognition and cleavage and to devise molecules that are not substrates of the standard proteasome to avoid killing healthy cells.
|
Traditional Medicinal Chemistry:
|
Synthesis of one-bead-one-compound libraries are generated and then screened for binding to a compontent of the 19S RP. It is the 19S RP that is responsible for recongizing, unwinding, and shuttling proteins into the catalytic core of the proteasome for degradation. Primary screening hits are tested in vivo for the desired biological activity. After this validation assay, the peptoid or peptide hits are used as probe molecules to screen for more drug-like candidates. Inhibitors of the 19S RP are highly effective in blood cancers.
|
Proudly powered by Weebly